INFORM
Discover Biological Insights
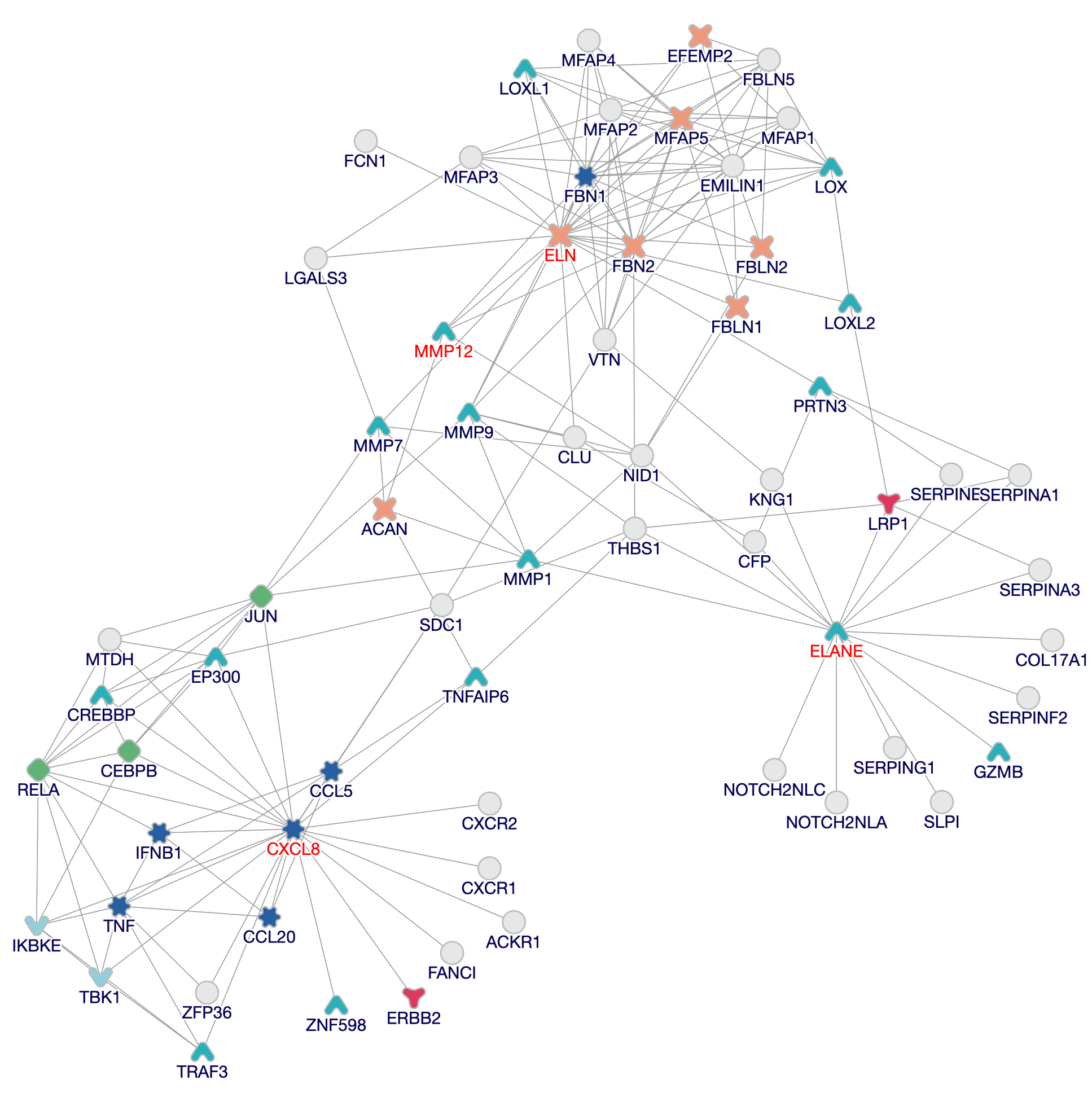
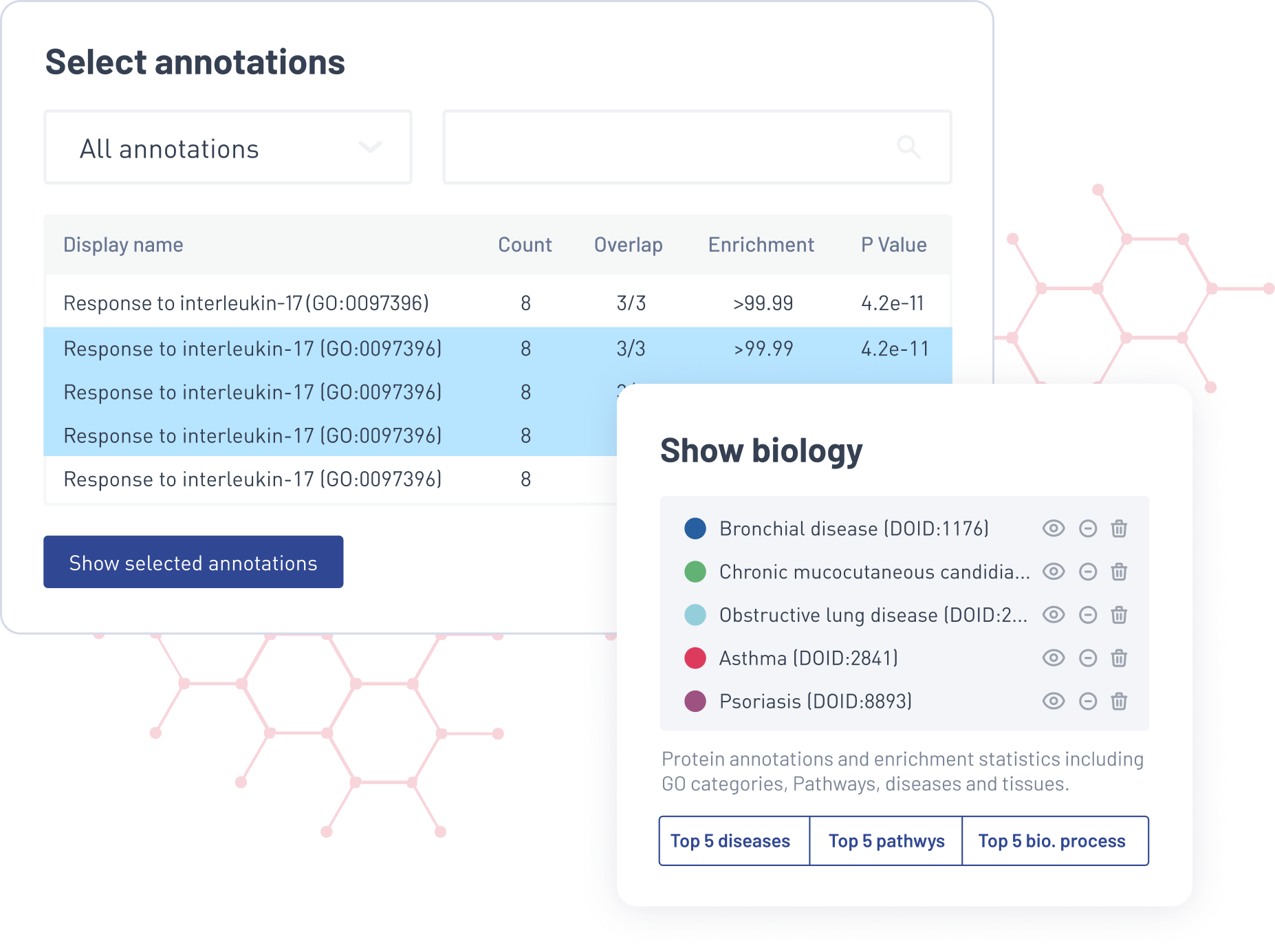
Explore a range of biological information. Make sense of your protein network with customizable annotations and visual cues. Highlight relevant disease pathways and uncover novel protein–protein interactions. Pinpoint which genes are statistically enriched across an array of biological processes.
INSPECT
Apply Your Data
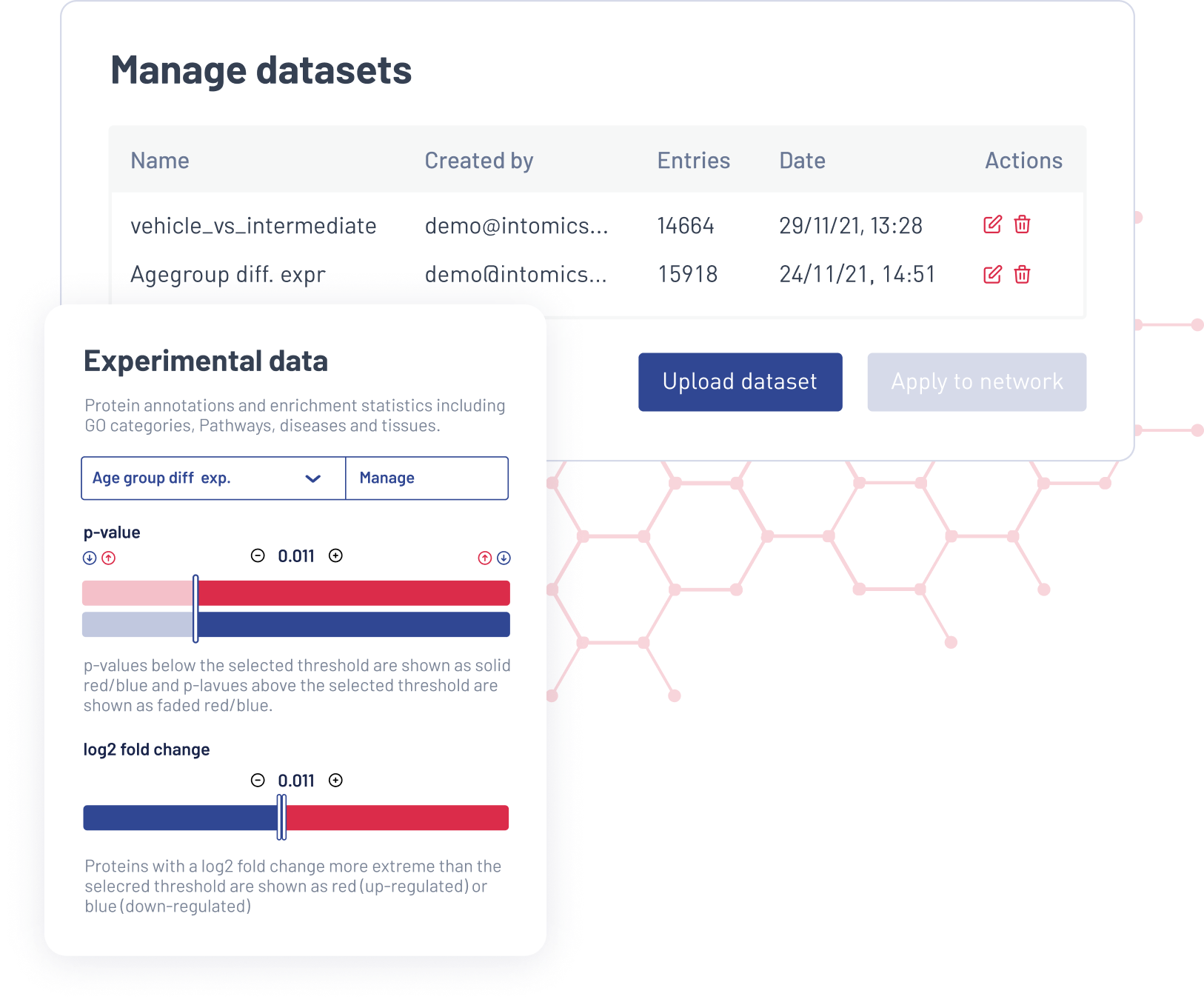
Integrate your data to unlock deeper network insights. Expose differential gene expression with interactive visualisations. Store your data and easily switch between data sets. Save your analysis and visualisations for further inspection.

INTERFACE
Customize Your Environment
Tailor a project workspace and save your relevant networks. Apply views and annotations across projects. Easily share your findings and enable further discovery.
Pricing Plans
Register for a free account, to get access to more features or get in contact to learn more about commercial licence and subscription plans.
Guest
The Interactome
Core Interactome
Protein information
Biological annotations
Fast enrichment analysis
Export network reports
5 protein interaction searches
3 annotation visualisations
Registered user Free
All guest features
Including the following upgrades:
Publish networks
1 experimental dataset
5 networks in workspace
5 protein interaction searches
5 annotation visualisations
2 custom annotations
License
All registered user features
Including the following upgrades:
Protein text mining
Export PPI information for networks up to 1000 proteins
Access to interaction evidence
10 experimental datasets
500 protein interaction searches
1000 networks in workspace
Unlimited annotation visualisations
Unlimited custom annotations
The Interactome
Protein Interaction Database
The Interactome database of high confidence protein-protein interactions is an ideal resource for elucidation of biological processes involved in diseases and to aid drug development.
- High-coverage map of human protein biology
- Highly trustworthy interactions extracted by scoring +6M traceable entries of interaction evidence
All publications using ZS Revelen or The Interactome data must cite Li, Wernersson et al., Nat. Methods 2017.
Data Sources
The following publications describe important data resources used for ZS Revelen and The Interactome
Nucleic Acids Res. 2003 Jan 1;31(1):248-50
BIND: the Biomolecular Interaction Network Database
Bader GD, Betel D, Hogue CW
Nucleic Acids Res. 2016 Jan 4; 44(D1):D7-19. doi: 10.1093/nar/gkv1290. Epub 2015 Nov 28
Database resources of the National Center for Biotechnology Information
NCBI Resource Coordinators
Nucleic Acids Res. 2009 Jan;37(Database issue):D5-15. doi: 10.1093/nar/gkn741. Epub 2008 Oct 21
Database resources of the National Center for Biotechnology Information
Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, Feolo M, Geer LY, Helmberg W, Kapustin Y, Landsman D, Lipman DJ, Madden TL, Maglott DR, Miller V, Mizrachi I, Ostell J, Pruitt KD, Schuler GD, Sequeira E, Sherry ST, Shumway M, Sirotkin K, Souvorov A, Starchenko G, Tatusova TA, Wagner L, Yaschenko E, Ye J
BMC Bioinformatics. 2009 Jun 16 ;10 Suppl 6:S3. doi: 10.1186/1471-2105-10-S6-S3
Databases of homologous gene families for comparative genomicsPenel S, Arigon AM, Dufayard JF, Sertier AS, Daubin V, Duret L, Gouy M, Perrière G
Nucleic Acids Res. 2016 Jan 4; 44(D1):D286-93. doi: 10.1093/nar/gkv1248. Epub 2015 Nov 17
eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences
Huerta-Cepas J, Szklarczyk D, Forslund K, Cook H, Heller D, Walter MC, Rattei T, Mende DR, Sunagawa S, Kuhn M, Jensen LJ, von Mering C, Bork P

Lottenborgvej 26, 2800 Kongens Lyngby / Denmark
- Cookie policy |
- Contact us |
- Terms of Service |
- Data Sources |
- Copyright © 2023 ZS Associates A/S |
- All rights reserved |